Contents
Module
23: Machine Learning Toolkit (App)
23.2.3. Lasso (least absolute shrinkage and selection operator)
23.2.7. Support vector machine (SVM Regression)
23.2.8. Neural Network Regression.
23.3.2. Nearest neighbors (KNN)
23.4. Self-organizing Map (SOM)
23.4.1. Unsupervised SOM (Clustering model)
23.4.2. Supervised SOM (Classification model)
23.4.3. Distributed Ensemble SOM (Advanced Classification SOM)
23.4.4. Ensemble Boosting SOM (Advanced Classification SOM)
Module 23: Machine Learning Toolkit (App)
Machine
Learning Toolkit (MLT) is a critical app in i2G data analytics
workflow. MLT on i2G offers the most advanced predictive algorithms including
both regression and classification. It also contains non-linear predictor
functions to model non-linear relationship between the input curves and the
target curve. Self-organizing Map is another advanced module built specifically
for facies classification by both supervised and unsupervised algorithms.
Key values:
o
Extraction of meaningful insights from
your data and information sources
o
Robust algorithms designed by
geoscientists and machine learning experts
o
Reproducible results by allowing seed
number input
o
Flexibility in configuring model inputs
and architectures
o
Easy-to-use and adjustable workflow
interface
o
Availability of different model
validation metrics: offer different ways to assess training and predicting
capacity: training/loss error/accuracy cross-plot as well as a confusion matrix
·
Machine Learning Toolkit on i2G
platform is an advanced tool that offers many state-of-the-art algorithms in
the industry. They are divided into 2 groups, namely Regression and
Classification.
·
Fundamentally, classification is about
predicting a label (discrete properties such as facies) and regression is about
predicting a quantity (continuous properties such as permeability).
·
Before running any ML technique, the
user is required to create a new ML
model.
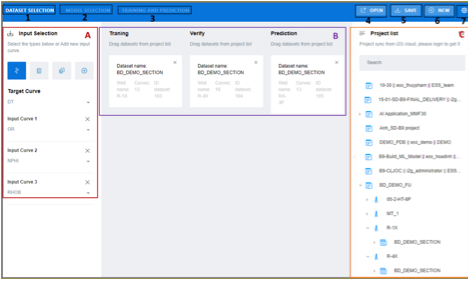
Figure 23‑1 Interface of Machine
Learning Toolkit app
The interface of machine learning app contains
three main regions: Tool Bar, Working
Area (A and B) and Project List
(C). The Tool Bar has 7 buttons
Table 23‑1 Buttons in Tool Bar
of MLT app
|
No |
Name |
Note |
|
1 |
“DATA SELECTION” tab |
Open the tab to select input data, including
input curves, target curve and input dataset for training/verify/ prediction |
|
2 |
“MODEL SELECTION” tab |
Open the tab to select machine learning
algorithms |
|
3 |
“TRAINING AND PREDICTION” tab |
Open the tab to execute the training,
validation and prediction process |
|
4 |
“OPEN” button |
Open other project(s) |
|
5 |
“SAVE” button |
Save the current project(s) |
|
6 |
“NEW” button |
Create a new project(s) |
|
7 |
Setting button |
Open a form for setting the text size |
Table 23‑2 Three separate spaces in the working areas
|
Area |
Note |
|
A |
Select input curves and the target curve |
|
B |
Dataset for training, verification and
prediction. These datasets should be dragged and dropped from project data
tree on the right (C) |
|
C |
List of projects. The user needs to select a
dataset, drop them to area B |
To build a machine learning model:
Step 1.
Go to https://ai.i2g.cloud/
Step 2.
Go to DATA
SELECTION tab > go to project list > select dataset for getting
training, verification and prediction data, then drag and drop them to area B
in the middle
Step 3.
Go to input selection area A on the left and select
one “Target” curve and input curves. The user can select them by curves name ![]() , curve
family
, curve
family ![]() or family group
or family group ![]() . Users
can click on the button (
. Users
can click on the button (![]() ) to add input curve.
) to add input curve.
Step 4.
Select MODEL
SELECTION tab > Select the type of model under “Type Selection”
> configure model architecture by adjusting model parameter
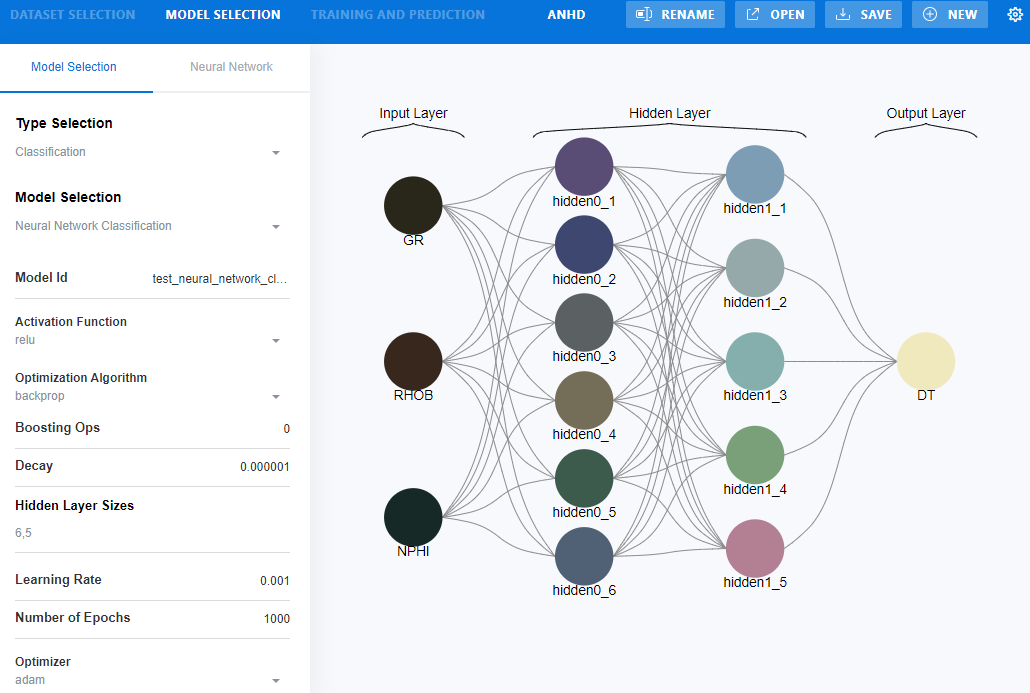
Figure 23‑2 configure model architecture in MLT
Step 5.
Select TRAINING
AND PREDICTION tab
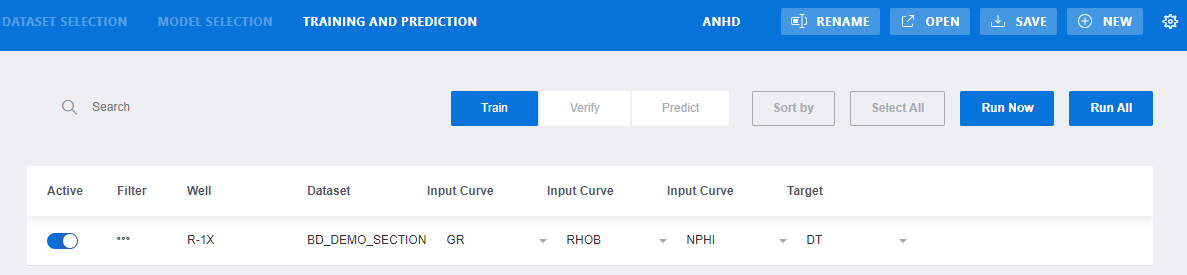
Figure 23‑3 Training and Prediction tab in MLT
Table 23‑3 Buttons in Training and Prediction tab in MLT
|
Button |
Note |
|
Active |
Activate/deactivate the selected dataset |
|
Filter |
Open a form to discriminate the input data |
|
Input Curve |
Select the input curve from the dataset |
|
Train |
Tab for configuring training data |
|
Verify |
Tab for configuring verifying data |
|
Predict |
Tab for configuring prediction data |
|
Run Now |
Run the current tab: Train or Verify or Predict |
|
Run All |
Run all tabs: Train/Verify/Predict |
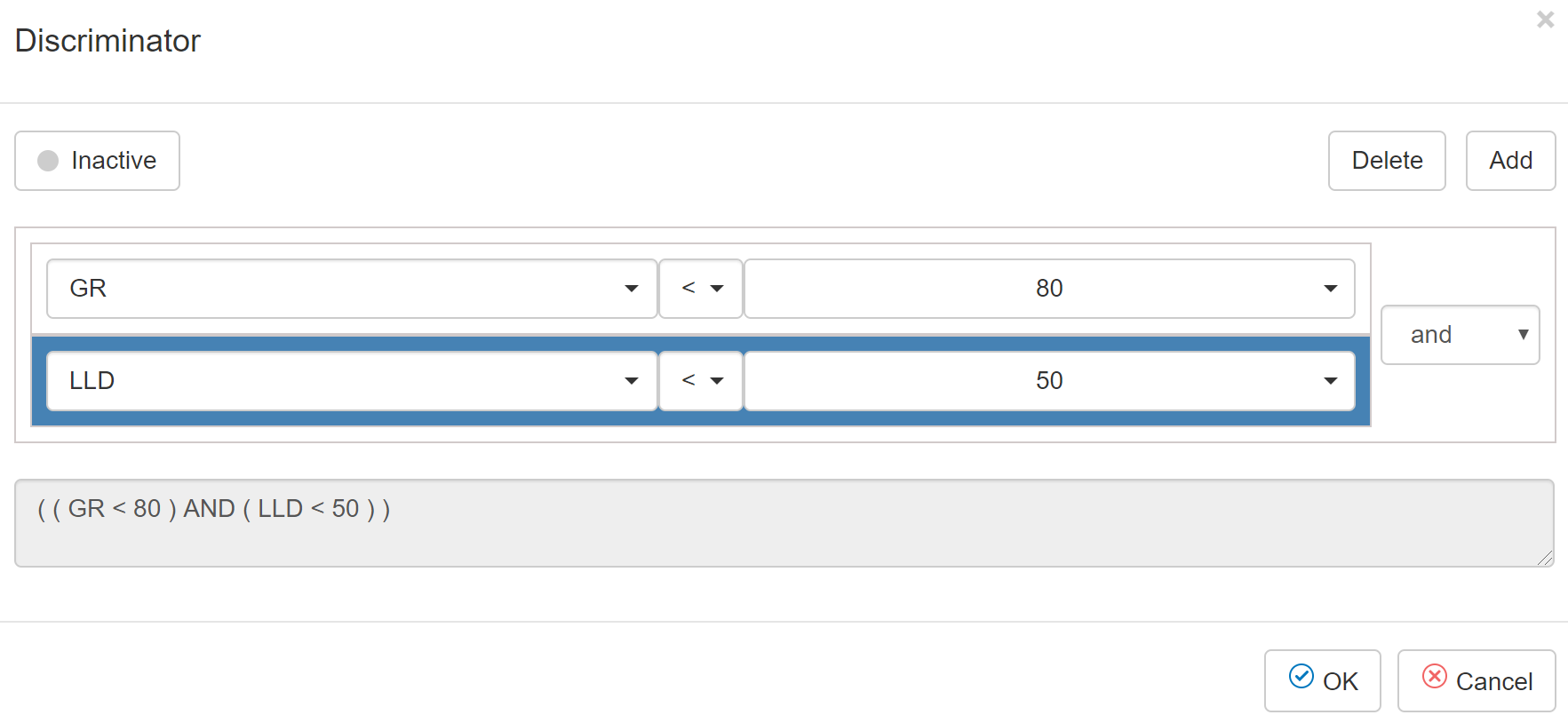
Figure 23‑4 The Discriminator dialog of the Training and
Prediction tab in MLT
Table 23‑4 Buttons in Discriminator dialog
|
Button |
Note |
|
Activated/inactive |
Select to activate/deactivate the filter |
|
Delete |
Delete the selected filter |
|
Add |
Add a filter. User can select either AND or OR |
To
create a filter before training a machine learning model:
Step 1.
Select filter icon ![]() in front of the dataset
in front of the dataset
Step 2.
Click this icon ![]() to set it to “Activated”
to set it to “Activated”
Step 3.
Select the curve used for discrimination > select
the comparison operators > input a value or select a curve to compare to
Step 4.
Click OK
This is a linear approach to modelling the relationship
between the input variables and output variable. The relationships are modeled
using linear predictor functions, so called linear models, whose unknown model
parameters are estimated from the data. Like all forms of regression analysis,
linear regression focuses on the conditional probability distribution of the outputs
given the values of the inputs, rather than on the joint probability
distribution of all of these variables, which is the domain of multivariate
analysis. All parameters of linear regression model are set as default and will
not be changed by the users.
Fit Intercept: (Boolean, default True) this parameter is used
to decide whether to calculate the intercept for this model. If set to False,
no intercept will be used in calculations (e.g. data is expected to be already
centered). In other words, the fitting line for the model is forced to pass the
origin (0, 0). If set to True, the line of best fit can "fit" the
y-axis. This means the line will not pass the origin.
Normalize: (Boolean, default True) this parameter is ignored
when the parameter Fit Intercept is set to False. If True, the inputs X will be
normalized to the range (-1, 1) before regression by subtracting the mean and
dividing by the standard deviation.
This is a regression method that finds the relationship
between inputs and outputs. Basically, that relationship is assumed to be a
model, which can be a mapping function, or some mapping rules, connecting the
two data. All parameters of the mapping model are ordinarily optimized based on
least squared error. This traditional approach will not work well if the
dataset is composed of outliers. In order to avoid the effect of outliers,
Huber loss function is introduced. Basically, Huber function is a penalty
function that is quadratic when the error is small, and is linear if the error
is large.
The input parameters are as follows.
Fit Intercept: (Boolean, default True) this parameter is used
to decide whether to calculate the intercept for this model. If set to False,
no intercept will be used in calculations (i.e. the data is already centered
around the origin.). In other words, the fitting curve for the model is forced
to pass the origin (0, 0). If set to True, the curve of best fit is allowed not
to pass the origin.
Tolerance: (real value, default 0.0001) is a threshold to
stop the training process. The iteration will stop if the maximum of projected
gradients with respect to all variables of the objective function is smaller
than Tol.
Alpha: (real value, default 1) this is the regularize
parameter, which is used to avoid overfitting. Increasing the value of alpha
results in less overfitting but also greater bias.
Epsilon: (real value greater than 1.0, default 2) this
parameter controls the number of samples that should be classified as outliers.
The smaller the epsilon, the more robust it is to outliers.
Max Number of Iterations: (integer, default 1000) this
parameter determines the maximum number of iterations that the fitting
algorithm should run for.
Warm Start: (Boolean, default True) This is useful if the
stored attributes of a previously used model has to be reused. If set to False,
then the coefficients will be rewritten for every call to fit.
23.2.3.
Lasso (least absolute shrinkage and selection operator)
This is a regression analysis method that performs both
variable selection and regularization in order to enhance the prediction
accuracy and interpretability of the statistical model it produces.
The input parameters for this model are as follows.
Random State: (integer) this parameter determines the seed of
the pseudo random number generator that selects a random feature to update.
Max Number of Iteration: (integer, default 1000) this is the
maximum number of iterations that the training process goes through.
Alpha: (real value, default 1) this is the coefficient
determining the relationship between output errors and the variance of the
inputs. Alpha = 0 is equivalent to an ordinary least square, solved by the
Linear Regression object. For numerical reasons, using Alpha = 0 with the Lasso
object is not advised. Given this, you should use the Linear Regression object.
Tolerance: (real value, default 0.0001) is the tolerance for
the optimization: if the updates are smaller than tolerance, the optimization
code checks the dual gap for optimality and continues until it is smaller than
Tolerance value.
This is a prediction model that uses a tree-like graph of
decisions and their possible consequences, including chance event outcomes,
resource costs, and utility. It breaks down a dataset into smaller and smaller
subsets while at the same time an associated decision tree is incrementally
developed. The final result is a tree with decision nodes and leaf nodes. A
decision node has two or more branches representing values for the attribute
tested. Each leaf node represents a decision on the numerical target. The
topmost decision node in a tree which corresponds to the best predictor is
called root node.
Input parameters for this model are as follows.
Max Depth of Trees: (integer, default 3) this parameter
determines the maximum depth of the tree. If “0”, then nodes are expanded until
all leaves are pure or until all leaves contain less than min_samples_split
samples.
Max Features: (integer, default 1) this is the number of
features to consider when looking for the best split.
Ranking Criterion: (integer, default “mse”) The function to
measure the quality of a split. Supported criteria are “mse” for the mean
squared error, which is equal to variance reduction as feature selection
criterion and minimizes the L2 loss using the mean of each terminal node,
“friedman_mse”, which uses mean squared error with Friedman’s improvement score
for potential splits, and “mae” for the mean absolute error, which minimizes
the L1 loss using the median of each terminal node.
Random State: (integer) this is the seed used by the random
number generator.
Splitter: (default “random”) The strategy used to choose the
split at each node. Supported strategies are “best” to choose the best split
and “random” to choose the best random split.
This is a regression method that constructs a multiple of
decision trees at training time and outputs the mean prediction of the
individual trees. This approach overcomes the overfitting behavior of single
decision trees. The collection of decision trees is generated such that each
individual tree is uncorrelated with the others by using different bootstrapped
samples from the training data.
The input parameters of the random forest regressor are as
follows.
Number of Estimators: (integer, default 150) is the number of
trees in the forest.
Max Depth: (integer, default 1) is the maximum depth of the
tree. If “0”, then nodes are expanded until all leaves are pure.
Max Features: the number of input features is used when
looking for the best split. The value of this parameter is the same as in
Decision tree.
Boostrap: (Boolean, default True) to determine whether
bootstrap samples are used when building trees. If False, the whole dataset is
used to build each tree.
Ranking Criterion: (default “mse”) The function to measure
the quality of a split. Supported criteria are “mse” for the mean squared
error, which is equal to variance reduction as feature selection criterion, and
“mae” for the mean absolute error.
Random State: is the seed used by the random number generator.
XGBoost stands for extreme gradient boosting method, which is
used for supervised learning problems, where we use the training data (with
multiple features) xi to predict a target variable yi. is an efficient and
scalable implementation of gradient boosting algorithm. Function estimation is
inspected from the aspect of numerical optimization in function space, rather
than parameter space. This is a more regularized model formalization to control
over-fitting, which gives it better performance than traditional gradient
boosting method.
Input parameters of this model are as follow.
Number of Estimators: (integer, default 150) number of trees
created.
Max Depth: (integer, default 5) Maximum depth of a tree.
Increasing this value will make the model more complex and more likely to
overfit. Beware that XGBoost aggressively consumes memory when training a deep
tree.
Gamma: (default 0.5) Minimum loss reduction required to make
a further partition on a leaf node of the tree. The larger gamma is, the more
conservative the algorithm will be. Range [0, ꝏ].
Booster: (Default “gbtree”) Which booster to use. Can be
gbtree, gblinear or dart; gbtree and dart use tree based models while gblinear
uses linear functions.
Regularization alpha: (integer, default 1) L1 regularization
term on weights. Increasing this value will make model more conservative.
Regularization lambda: (integer, default 1) L2 regularization
term on weights. Increasing this value will make model more conservative.
Random State: is the seed used by the random number
generator.
23.2.7.
Support vector machine (SVM Regression)
Support vector machine for regression works in a way that is
almost the same as for classification problems, except that the maxima margin
is added by a tolerance factor epsilon, which is set in approximation to the
SVM such that it can adapt to the real input values. Input parameters of SVM
regression model are as follows.
Kernel: (default “rbf”) name of kernel function
C: (default 2) this is the parameter for the soft margin cost
function, which controls the influence of each individual support vector. A
large C gives you low bias and high variance, which means you penalize the cost
of error a lot. A small C gives you higher bias and lower variance, which means
higher generalization.
Gamma: (default 0.1) kernel parameter for ‘rbf’, ‘poly’ and
‘sigmoid’. This parameter can affect the data transformation behavior.
Intuitively, a small gamma value means two points can be considered similar
even if are far from each other. In the other hand, a large gamma value means
two points are considered similar just if they are close to each other. So a
small gamma will give you low bias and high variance while a large gamma will
give you higher bias and low variance.
Degree: (default 2) is the degree of the polynomial kernel
function (‘poly’). Ignored by all other kernels.
Tolerance: (default 0.001) Tolerance for stopping criterion..
Max Number of Iterations: (default 100) Hard limit on
iterations within solver, or -1 for no limit.
23.2.8.
Neural Network Regression
It is a class of feedforward artificial neural network. A
neural network consists of three types of layers of nodes, including input
layer, hidden layers, and output layer. Except for the input nodes, each of
other nodes is a neuron that uses a nonlinear activation function. NN can be
used for both classification and regression problems. The supervised learning
technique utilized for NN is backpropagation algorithm. Input parameters for
this model are defined as follows.
Solver: (default “lbfgs”) The solver for weight optimization.
·
‘lbfgs’ is an optimizer in the family of quasi-Newton
methods.
·
‘sgd’ refers to stochastic gradient descent.
·
‘adam’ refers to a stochastic gradient-based optimizer
proposed by Kingma, Diederik, and Jimmy Ba
·
‘Cg’ is the conjugate-gradient optimization algorithm.
Apart from the well-known backpropagation algorithm, users can also use
Conjugate Gradient as a training algorithm for Neural networks. All input
parameters for conjugate gradient neural network have the same meaning as those
in MLP.
Note: The
default solver ‘adam’ works pretty well on relatively large datasets (with
thousands of training samples or more) in terms of both training time and
validation score. For small datasets, however, ‘lbfgs’ can converge faster and
perform better.
Activation Function: (default “relu”) is the name of
activation function for hidden nodes.
·
‘identity’, no-op activation, useful to implement
linear bottleneck, returns f(x) = x
·
‘logistic’, the
logistic sigmoid function, returns f(x) = 1 / (1 + exp(-x)).
·
‘tanh’, the
hyperbolic tan function, returns f(x) = tanh(x).
·
‘relu’, the
rectified linear unit function, returns f(x) = max(0, x)
Max number of iterations: (integer, default 500) is the
maximum number of iterations. The solver iterates until convergence (determined
by the parameter ‘tolerance’) or this number of iterations. For stochastic
solvers (‘sgd’, ‘adam’), note that this determines the number of epochs (how
many times each data point will be used), not the number of gradient steps.
Tolerance: (default 0.0003) Tolerance for the optimization.
When the loss or score is not improving by at least Tol for two consecutive
iterations, convergence is considered to be reached and training stops.
Learning rate initialization: (default 0.001) It controls the
step-size in updating the weights. Only used when solver is ’sgd’ or ‘adam’.
This is a non-parametric supervised learning method used for
classification or regression. The goal is to create a model that predicts the
value of a target variable by learning simple decision rules inferred from the
data features. The decision tree is built by recursively selecting the best
attribute to split the data and expanding the leaf nodes of the tree until the
stopping cirterion is met. The choice of best split test condition is
determined by comparing the impurity of child nodes and also depends on which
impurity measurement is used. If the decision trees are too large, overfitting
may occur. Input parameters for decision tree classifiers are defined as
follows.
Ranking Criterion: (default “entropy”) is the function to
measure the quality of split.
·
“gini”: is for the Gini impurity. This is a measure of
how often a randomly chosen element from the set would be incorrectly labeled
if it was randomly labeled according to the distribution of labels in the
subset. It reaches its minimum (zero) when all cases in the node fall into a
single target category.
·
“entropy”: is for the information gain. Information
gain is used to decide which feature to split on at each step in building the
tree. In order to keep the tree small, at each step we should choose the split
that results in the purest daughter nodes, which correspond to the feature with
highest information gain with the output labels. The split with the highest
information gain will be taken as the first split and the process will continue
until all children nodes are pure, or until the information gain is 0.
Min samples split: (integer, default 5) is the minimum number
of samples required to split an internal node. The larger the min_samples is,
the bigger trees can be created.
Min Impurity Decreases: (Real values, default 0.01) is the
threshold for early stopping in tree growth. A node will split if its impurity
is above the threshold, otherwise it is a leaf. This value will also determine
the size of trees.
23.3.2.
Nearest neighbors (KNN)
This is a non-parametric method used for classification. The
input consists of the k closest training examples in the feature space. The
output is a class membership. An object is classified by a majority vote of its
neighbors, with the object being assigned to the class most common among its K
nearest neighbors (K is a positive integer, typically small). If K = 1, then
the object is simply assigned to the class of that single nearest neighbor.
Input parameters for this model is defined as follows.
Number of Neighbors: (default 100) is the number of closest
training samples to the testing sample.
P: (default 1) is the power parameter for the Minkowski
metric. It determines how to calculate the distance between samples. When p =
1, this is equivalent to using manhattan_distance (l1), and euclidean_distance
(l2) for p = 2. For arbitrary p, minkowski_distance (l_p) is used.
Distance Metric: (minkowski distance) determines the method
to calculate the distance between samples
This classification algorithm is named for the function used
at the core of the method, the logistic function. The logistic function, also
called the sigmoid function is an S-shaped curve that can take any real-valued
number and map it into a value between 0 and 1, but never exactly at those limits.
f = 1 / (1 + e^-value), where e is the base of the natural logarithms. There
can be many different equations used for logistic regression, for example: y =
e^(b0 + b1*x) / (1 + e^(b0 + b1*x)), where y is the predicted output, b0 is the
bias or intercept term and b1 is the coefficient for the single input value
(x). Each column in your input data has an associated b coefficient (a constant
real value) that must be learned from your training data.
Logistic regression is a linear method, but the predictions
are transformed using the logistic function. The impact of this is that we can
no longer understand the predictions as a linear combination of the inputs as
we can with linear regression.
Logistic regression models the probability of the default
class. In this model we use 30 logistic regression classifiers corresponding to
30 different default classes. For each classifier, the default class is fitted
against all the other classes, resulting in the probability of the data sample
being 1 of the 30 classes. In the prediction phase, the class with the highest
output probability will be assigned to the testing sample.
The setting parameters for this model are described as
follows.
C: (default 20) is the inverse of regularization strength.
Like in support vector machines, smaller values specify stronger
regularization.
Max Number of Iterations: (default 10000) Maximum number of
iterations taken for the solvers to converge.
Solver: (default “liblinear”) is the name of the algorithm to
use in the optimization problem.
A random forest is a meta estimator that fits a number of
decision tree classifiers on various sub-samples of the dataset and use
averaging to improve the predictive accuracy and control over-fitting. The
sub-sample size is always the same as the original input sample size but the
samples are drawn with replacement if bootstrap=True (default).
Input parameters for this method are defined as follows.
Number of Trees: (default 150) The number of decision trees
in the forest
Ranking Criterion: (default “entropy”) The function to
measure the quality of a split. This is the same as in decision tree.
Min Samples Split: (default 5) The minimum number of samples
required to split an internal node. This is the same as in decision tree.
Min Impurity Decrease: (default 0.0003) Threshold for early
stopping in tree growth. This is the same as in decision tree.
This is a multilayer perceptron network for classification
problem. This model optimizes the log-loss function using LBFGS or stochastic
gradient descent. Input parameters for this model are defined as follows.
Activation Function: (default “relu”) is the name of
activation function for hidden layers.
Optimization Algorithm: (default “backprop”) is the name of
training algorithm, which is either backpropagation or evolution strategy
algorithm.
Backpropagation algorithm works best with a large number of
epochs.
Evolution algorithm is preferable with smaller number of
epochs, about 1000.
Learning Rate: (default 0.001) It controls the step-size in
updating the weights.
Number of Epochs: (default 1000) maximum number of training
iterations.
Optimizer: (default “adam”) is the solver for weight
optimization.
Warm up: (default “false”) is to determine the initial values
of weights. When set to True, the model will run with batch_size = 1 and
num_epochs = 1 to get the initial weights, otherwise, it just randomizes the
initial weights. After this process, the model will run the training process as
set by other parameters.
Boosting Ops: (default 0) this parameter is only used for
evolution algorithm. This parameter determines the number of times that the
weights are further updated based on the accuracy after the training process
based on the loss function finishes. Default is 100.
Sigma: (default 0.01) this parameter is only used for
evolution algorithm. This parameter determines the variance size of the
Gaussian distribution used for randomizing the weights.
Population: (default 50) is the number of sets of randomized
weights for each epoch. This parameter is only used for evolution algorithm.
Decay: (default 0.000001) is to determine the decaying rate
of weight updates.
23.4.
Self-organizing Map (SOM)
Self-Organizing Maps (SOMs) is a type of artificial neural
network that is trained using unsupervised learning to produce a
low-dimensional (typically two-dimensional), discretized representation of the
input space of the training samples, called a map, and is therefore a method to
do dimensionality reduction. Self-organizing maps apply competitive learning in
the sense that they use a neighborhood function to preserve the topological
properties of the input space. This makes SOMs useful for visualization by
creating low-dimensional views of high-dimensional data.
SOM can provide an interpretation of the dataset about data
distribution, the number of clusters and some other visualizations. Thus, SOM
is traditionally considered as both a dimensional reduction tool and a
clustering tool. It has been shown that while self-organizing maps with a small
number of nodes behave in a way that is similar to the K-means algorithm,
larger self-organizing maps rearrange data in a way that is fundamentally
topological in character.
In general, the weights of the neurons are initialized either
to small random values, or by randomly choosing samples from the training data
to initialize the weights of neurons, or sampled evenly on the plane formed by
the two largest principal component eigenvectors.
The self-organizing system in SOM is a set of nodes (or
neurons) connected to each other via a topology of rectangle. This set of nodes
is called a map. Each neuron has several nearest neighbors (4 or 8 with
rectangular topology). At each training step, an input vector Xi is introduced
to the map, using the Euclidean distance, the different between Xi and each
neuron in the map is calculated. The most similar neuron to the selected sample
is called the winning node or the best-matching unit (BMU). The weight vector
of the BMU is then updated by a learning rule:
W_i (t+1)= W_i (t)+
θ(t).α(t).(X_i - W_i (t))
Where W_i (t+1) is the newly updated weight vector of BMU at
time t+1. α(t) is the learning rate which decreases over each training
iteration. By this learning rule, the BMU is moved closer to the chosen input
vector. Instead of applying the winner-takes-all principal, the BMU’s
neighboring neurons can also be updated. The neighborhood of BMU is determined
by BMU’s radius. The learning rate of BMU, α(t), is decaying after each
updating iteration as follows:
α(t)=α_0/(1+ decay_rate *t)
Where, t is the maximum number of update iterations; α_0 is
the initial learning rate.
θ(t) is the neighboring index determining how much a
neighboring neuron is updated in each iteration. Depending on different
neighboring methods, θ(t) can be calculated differently as:
·
Bubble method: all neurons inside the distance from
the BMU will be updated.
θ(t)={■(1 if dist≤σ(t)@0 if
dist>σ(t))┤
Where, σ(t) is the distance function between BMU and other
neurons, which also decays during the update process.
·
Gaussian method: all neurons will be updated with the
distance index defined as
θ(t)=e^(-〖dist〗^2/(2〖σ(t)〗^2 ))
Where, “dist” is the Euclidean distance between the BMU and
other neurons.
tIn those methods, The distance function σ(t) is defined as
σ(t)=σ_0/(1+sigma_decay_rate *t)
Where σ_0 is the initial neighboring radius, and the
sigma_decay_rate determines how much the distance function decays after each
iteration.
23.4.1.
Unsupervised SOM (Clustering model)
This is a traditional use of the SOM as discussed above. The
input data samples are clustered using SOM. Users can manage following
parameters:
Number of Columns: (integer, default 10) This is the number
of columns of neurons in the topology of rectangle of the SOM.
Number of Rows: (integer, default 10) This is the number of
rows of neurons in the topology of rectangle of the SOM.
Random State: (integer) this is the seed used by the random
number generator.
Learning Decay Rate: (real) is the decay_rate.
Learning Rate: (real, in range (0, 1)) is the initial
learning rate α_0.
Neighborhood: (bubble or Gaussian) is the method to determine
the neighboring index θ(t).
Weight Initialization: (random, sample, or PCA) is the method
to initialize the weights of all neurons in the SOM.
Random: the weights are generated as small random values,
Sample: the weights are generated by randomly choosing
samples from the training data
PCA: the weights are sampled evenly on the plane formed by
the two largest principal component eigenvectors of the training data.
Sigma: (real) is the initial neighboring radius, σ_0.
Sigma Decay Rate: (real) is the rate for the neighboring
radius to decay over updating iterations.
Batch Size: (integer) is the number of samples in the same
batch during learning phase.
Number of Clusters: (integer) is the number of clusters that
users want to divide the data using SOM.
23.4.2.
Supervised SOM (Classification model)
This is a combination method between the traditional SOM and
the Learning Vector Quantization (LVQ) algorithm to form a classification
version of SOM. SOM will be supervised trained using labeled training data to
create a classification model.
The supervised training process for SSOM includes two phases
as depicted in the below figure
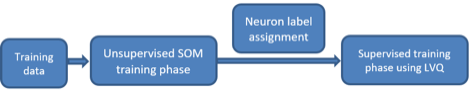
Figure 23‑5 Two phases of the supervised training process for
SSOM
Unsupervised SOM training phase follows traditional SOM
algorithm. The input training dataset is used to generate the clustering model
of the SOM.
After the first phase, all neurons in the SOM are labeled
based on the closest training samples.
Supervised training phase is based on LVQ algorithm, which
moves the labeled neurons closer to the training samples having the same
labels, or moves the neurons further to the sample having different labels.
Users can manage following parameters:
Number of Columns: (integer, default 10) This is the number
of columns of neurons in the topology of rectangle of the SOM.
Number of Rows: (integer, default 10) This is the number of
rows of neurons in the topology of rectangle of the SOM.
Random State: (integer) this is the seed used by the random
number generator.
Learning Decay Rate: (real) is the decay_rate.
Learning Rate: (real, in range (0, 1)) is the initial learning
rate α_0.
Neighborhood: (bubble or Gaussian) is the method to determine
the neighboring index θ(t).
Weight Initialization: (random, sample, or PCA) is the method
to initialize the weights of all neurons in the SOM.
Random: the weights are generated as small random values,
Sample: the weights are generated by randomly choosing
samples from the training data
PCA: the weights are sampled evenly on the plane formed by
the two largest principal component eigenvectors of the training data.
Sigma: (real) is the initial neighboring radius, σ_0.
Sigma Decay Rate: (real) is the rate for the neighboring
radius to decay over updating iterations.
Sup Batch Size: (integer) is the number of samples in the
same batch during supervised learning phase.
Sup Number of Iterations: (integer) is maximum number of
supervised training iterations.
Sup Batch Size: (integer) is the number of samples in the
same batch during supervised learning phase.
Sup Number of Iterations: (integer) is maximum number of
supervised training iterations.
Unsup Batch Size: (integer) is the number of samples in the
same batch during unsupervised learning phase.
Unsup Number of Iterations: (integer) is maximum number of
unsupervised training iterations.
23.4.3.
Distributed Ensemble SOM (Advanced Classification SOM)
This is an improved classification model based on supervised
SOM. In this model, multiple SOM ensembles are generated based on multiple
subsets of input features. Each feature subset is selected either by random or
by weighting method. One feature subset is used to create a classification SOM
model. Since the size of feature subset is much smaller than the original
feature set, the complexity of each ensemble SOM structure can be reduced.
Moreover, if the original data is divided into multiple subsets with different
feature sets, the information of the training data can be closer analyzed under
different angles. This results in a better performance of the classification
algorithm. Also, the combination of multiple ensemble SOM may help reduce the variance
of the classification errors. From a set of local decisions provided my
multiple ensemble SOMs, the test sample is assigned a label based on the
majority voting rule.
In this method, the training feature set is divided into
multiple feature subset. This looks like a system managed by multiple
distributed sensors, each of which contributes a different set of measurements.
The name of this method is motivated from this implication.
In this method, users can manage following parameters:
Number of Estimator: (integer, default 30) is the number of
ensemble SOMs created in the algorithm.
Random State: (integer) this is the seed used by the random
number generator.
Size of Estimator: (integer, default 4) is the size of the
ensemble SOMs with a square topology. Here, “Size” is the number of rows as
well as the number of columns of each ensemble SOM.
Learning Decay Rate: (real) is the decay_rate.
Learning Rate: (real) is the initial learning rate α_0.
Neighborhood: (bubble or Gaussian) is the method to determine
the neighboring index θ(t).
Sigma: (real) is the initial neighboring radius, σ_0.
Sigma Decay Rate: (real) is the rate for the neighboring
radius to decay over updating iterations.
Sup Batch Size: (integer) is the number of samples in the
same batch during supervised learning phase.
Sup Number of Iterations: (integer) is maximum number of
supervised training iterations.
Subset Size: (real, in the range (0, 1]) is the size of
training data selected for SOM learning processes.
Sup Number of Iterations: (integer) is maximum number of
supervised training iterations.
Unsup Batch Size: (integer) is the number of samples in the
same batch during unsupervised learning phase.
Unsup Number of Iterations: (integer) is maximum number of
unsupervised training iterations.
Feature Selection: (random or weights) is the method to
generate feature subsets
Random: each feature subset is generated by uniformly
selection from a pool of original feature set.
Weights: the features are first assigned weights according to
their correlation coefficients with the labels. Each feature set is made from a
weighted selection from the original feature set. Features with higher weights
tend to be selected more often into feature subsets than the ones with smaller
weights.
23.4.4.
Ensemble Boosting SOM (Advanced Classification SOM)
Motivated from boosting algorithm using multiple ensembles.
This approach aims at creating multiple simple SOM models, each of which is
created from a subset of training data. In general, each training subset is randomly
selected from the training dataset. Each training subset is used to train a
supervised SOM model. Decisions from multiple SOMs will be fused based on the
majority voting rule. This approach may produce a better performance than
single supervised SOM model since it helps reduce the variance of the
classification errors.
In this method, users can manage following parameters:
Number of Estimator: (integer, default 50) is the number of
ensemble SOMs created in the algorithm.
Random State: (integer) this is the seed used by the random
number generator.
Size of Estimator: (integer, default 4) is the size of the
ensemble SOMs with a square topology. Here, “Size” is the number of rows as
well as the number of columns of each ensemble SOM.
Weight Initialization: (random, sample, or PCA) is the method
to initialize the weights of all neurons in the SOM.
Random: the weights are generated as small random values,
Sample: the weights are generated by randomly choosing
samples from the training data
PCA: the weights are sampled evenly on the plane formed by
the two largest principal component eigenvectors of the training data.
Learning Decay Rate: (real) is the decay_rate of the learning
rate.
Learning Rate: (real) is the initial learning rate α_0.
Neighborhood: (bubble or Gaussian) is the method to determine
the neighboring index θ(t).
Sigma: (real) is the initial neighboring radius, σ_0.
Sigma Decay Rate: (real) is the rate for the neighboring
radius to decay over updating iterations.
Sup Batch Size: (integer) is the number of samples in the
same batch during supervised learning phase.
Sup Number of Iterations: (integer) is maximum number of
supervised training iterations.
Subset Size: (real, in the range (0, 1]) is the size of
training data selected for SOM learning processes.
Sup Number of Iterations: (integer) is maximum number of
supervised training iterations.
Unsup Batch Size: (integer) is the number of samples in the
same batch during unsupervised learning phase.
Unsup Number of Iterations: (integer) is maximum number of
unsupervised training iterations.
This is a nonlinear approach to model the relationship
between the input curves and the target curve. The relationships are modeled
using nonlinear predictor functions, so called nonlinear models, whose unknown
model parameters are estimated from the data. The nonlinear relationship
function is defined by the user, where the variables x1, x2, … correspond to
the input curves from top to bottom, respectively.
Table 23‑5 Guides to input nonlinear functions
|
Algebra expression |
Input Syntax |
Meaning |
|
a . x |
a*x |
a multiplied by x |
|
a : x |
a/x |
a divided by x |
|
a + x |
a+x |
a plus x |
|
a - x |
a-x |
a minus x |
|
|
a**x |
a power x |
|
Ln(x) |
log(x) |
Natural logarithm of x |
|
Log(x) |
log(x)/log(10) |
Base 10 logarithm of x |
|
Loga(x) |
log(x)/log(a) |
Base a logarithm of x |
|
e |
e |
Natural number (2.71828). No other
parameter named e is permitted. |